Unraveling the mechanisms that create the individualized metabolism in leukemia

A research collaboration based in Kumamoto University (Japan) has shown that lysine-specific demethylase 1 (LSD1), an enzyme involved in gene expression, produces individualized metabolism depending on the type of acute myeloid leukemia cells. Cancer cells are known to have a unique ability to metabolize substances differently from normal cells, and this ability is considered to be a promising therapeutic target. The new research findings may contribute to the safe and effective use of LSD1 inhibitors as potential anticancer agents, and to the development of highly specific treatments for various leukemia types.Acute myelogenous leukemia (AML) occurs when hematopoietic stem cells become tumors rather than differentiating into white or red blood cells. The various types of AML develop according to which stage of differentiation they become cancerous. Those that develop when differentiating into red blood cells are classified as erythroblastic leukemia (EL). Although pathology-specific targeted molecular therapies have been developed for some forms of AML, and have improved treatment outcomes, many forms of the disease, including EL, have high mortality rates due to the lack of individualized therapies. Thus, therapies based on disease type and molecular pathology are desired.
Recent studies have revealed that the inherent metabolic capacity of cancer cells contributes significantly to tumor formation, metastasis and resistance to treatment. Therefore, therapeutic strategies that target nutrient transport and metabolic pathways active in cancer cells have been devised, but it has also been pointed out that metabolic characteristics vary depending on the type and progression of cancer. The metabolic characteristics of AML have not been fully investigated and, in particular, the differences that depend on the disease type and their mechanisms have not been clarified.
Gene expression is regulated by the epigenome. Chemical modifications, such as DNA methylation and methylation of the histone proteins around which the DNA wraps, act as markers to shape the epigenome. There are many differences in the epigenome between cancer cells and normal cells, which in turn generate the differences in gene expression patterns.
Previously, the Kumamoto University research collaboration showed that LSD1, a demethylase that removes methyl groups from methylated histones, is involved in the regulation of energy metabolism in various cell types (Nature Communications 2012, Cancer Research 2015, Nucleic Acids Research 2018). Therefore, they decided to test the possibility that LSD1 is involved in the metabolic regulation of AML cells. Although it has been shown that LSD1 inhibitors may be effective in the treatment of AML, not much is known about the differences in efficacy depending on the disease type. Thus, in this study, the researchers decided to focus on the metabolic differences depending on AML disease type and the role of LSD1.
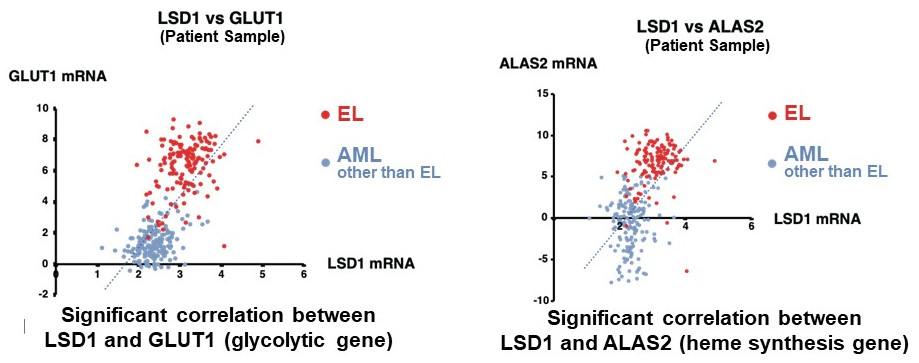
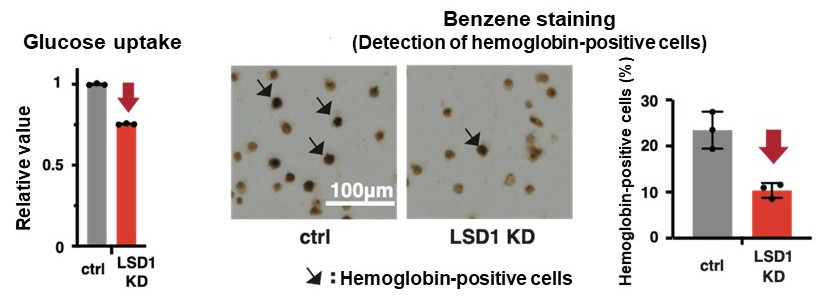
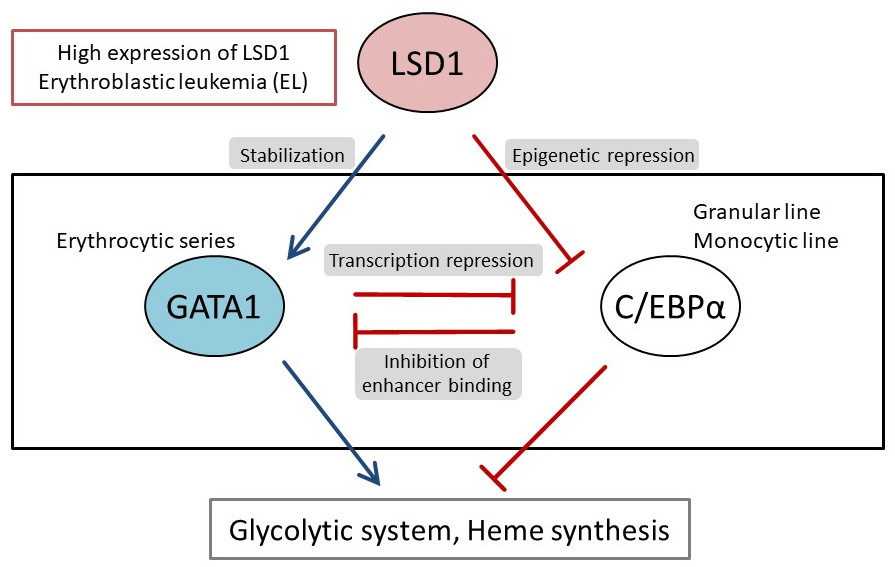
They first analyzed the gene expression database of AML patients and AML-derived cultured cell lines, and found that both LSD1 and glycolytic genes are highly expressed in EL among AML patients. Then, when they tested the inhibition of LSD1 function using EL cell lines, they found that LSD1 promoted glucose uptake into cells and the glycolytic system. An integrated omics analysis revealed that heme synthesis, a characteristic metabolic pathway of normal red blood cells, is also activated by LSD1. The mechanism was found to be that LSD1 activates glycolytic and heme synthesis gene expression by preventing the degradation of GATA1 protein, an erythroid transcription factor.
Furthermore, under LSD1 functional inhibition, the expression of CEBP/α, a transcription factor of the granulocyte-monocyte lineage of leukocytes, was dramatically up-regulated, inhibiting metabolic regulation by GATA1. These results indicate that LSD1 regulates the balance of transcription factors involved in hematopoietic cell lineage, thereby generating the metabolic phenotype characteristic of EL. Additionally, analysis of clinical data covering various forms of AML showed that the expression of LSD1, GATA1, and glycolytic and heme synthesis genes showed a significant positive correlation. This suggests that regulation of cellular lineage by LSD1 may generate the diversity of metabolic types of AML.
"Based on our research, a combination of LSD1 inhibitors, metabolic target drugs, and conventional therapies may be highly effective in treating patients with EL who express high levels of LSD1," said Associate Professor Shinjiro Hino, who led the study. "It may also be an important clue for selecting patients who may benefit from LSD1 inhibitors in clinical trials."
This research was posted online in Blood Advances on 30 April 2021.
[Source]
Kohrogi, K., Hino, S., Sakamoto, A., Anan, K., Takase, R., Araki, H., ... Nakao, M. (2021). LSD1 defines erythroleukemia metabolism by controlling the lineage-specific transcription factors GATA1 and C/EBPα. Blood Advances, 5(9), 2305–2318. doi:10.1182/bloodadvances.2020003521
[Funding]
- KAKENHI Grants from MEXT Japan (JP18H02618, 18K19479, 20H04108, JP16K07215, 25430178, 20K16335),
- Takeda Science Foundation,
- The SGH Foundation,
- The Japan Agency for Medical Research and Development (JP16gm0510007)
[Usage Restrictions]
Image credits belong to Associate Professor Shinjiro Hino unless otherwise indicated in the figure caption. To use any of the media contained within this release elsewhere, a reference to the original work (this release) should be included and other restrictions must be followed as indicated.