Amygdala changes in male patients with schizophrenia and bipolar disorder
DNA hypermethylation responsible for reduced amygdala volume in male patients with schizophrenia and bipolar disorder.
Researchers at Kumamoto University, Japan have revealed that DNA methylation occurs in the gene that codes serotonin transporter (SERT), a protein that regulates neurotransmitter transmission, in schizophrenia and bipolar patients. Particularly prominent in males and patients with certain genetic polymorphisms, this methylation is inversely correlated with volume of the amygdala in the brain. This work is expected to lead to a better understanding of the pathophysiology of schizophrenia and bipolar disorder. It may also help to develop therapeutic agents and diagnostic/therapeutic markers that target epigenetic conditions.
Schizophrenia and bipolar disorder are serious mental illnesses that affect approximately 1% of the population and require long-term treatment. Past epidemiological studies suggest that onset involves a complex interplay of genetic and environmental factors, but no definitive genetic factors have been identified yet.
SERT regulates the concentration of the neurotransmitter serotonin within the gap between neurons where neurotransmitters are exchanged (the synaptic cleft). It is considered to be deeply involved with mental illnesses, and drugs targeting the protein are widely used as treatments for depression and anxiety disorders. In particular, a polymorphism called 5-HTTLPR in the gene that encodes SERT has been a target for many mental disorder studies. When the polymorphism is the L (long) type, more SERT is produced, and when it is the S (short) type, less is produced. People generally have one combination of these types, L/L, S/L, or S/S. In 2003, an analysis (Capsi et al., 2003) became highly regarded for reporting that people with the S-type polymorphism had a stronger tendency for anxiety and were more susceptible to depression. However, recent large-scale studies scrutinized that work and clearly denied the simple association between the 5-HTTLPR polymorphism and mental illness.
Expression of genes in DNA is controlled by chemical modifications like methylation. A collaborative research group between Kumamoto University and the University of Tokyo used DNA methylation analysis in patients with bipolar disorder (Sugawara et al., 2011) to show that two specific sites (CpG3 and CpG4) in the SERT gene show high methylation status. Here, they conducted a large-scale follow-up study on those CpG sites, a new study with patients having schizophrenia, and a comprehensive study on the pathology of SERT DNA methylation.
They measured the DNA methylation status of the SERT gene using genomic DNA extracted from the blood of 450 patients with bipolar disorder, 440 patients with schizophrenia, and 460 healthy subjects. They found that the SERT CpG3 site in patients with bipolar disorder and schizophrenia is hypermethylated in male patients. Additionally, after administration of an antipsychotic drug over four weeks, no change in methylation was detected in animal test subjects (marmoset, n=3) compared to control (n=3), which suggests that methylation in bipolar and schizophrenic patients is not affect by medication.
Furthermore, a detailed analysis of the SERT gene polymorphism 5-HTTLPR revealed that the methylation rate was high in patients with bipolar disorder and schizophrenia when 5-HTTLPR was hypoactive type (S/S or S/L). When the CpG3 site of the SERT gene was artificially methylated, researchers found that the transcriptional activation ability and production of the SERT protein were both markedly suppressed.
Researchers also investigated the volume of the amygdala with MRI images from 41 healthy subjects and 57 schizophrenia age- and gender-matched patients. The amygdala is a part of the brain that, among other functions, plays a major role in emotional responses. SERT actions are strong there and its association with DNA methylation has been reported in the past. The volume of the left amygdala of male patients with hypoactive type 5-HTTLPR polymorphism was found to have an inverse correlation with the DNA methylation rate of CpG3. This suggests that male schizophrenia patients of this polymorphism type may have a hypermethylated SERT gene that leads to decreased amygdala volume through decreased SERT protein levels.
"Our research shows that a specific site in the SERT gene is hypermethylated in male patients with schizophrenia and bipolar disorder, and is associated with volume changes in the amygdala," said study leader, Dr. Kazuya Iwamoto. "In the future, we believe that this can be used as a biomarker for onset prediction, diagnoses, and therapy assessments. It could even be a clue for the molecular pathologies of these disorders."
This research was posted online in Schizophrenia Bulletin on 19th June 2020.
[Source]
Ikegame, T., Bundo, M., Okada, N., Murata, Y., Koike, S., Sugawara, H., … Iwamoto, K. (2020). Promoter Activity-Based Case-Control Association Study on SLC6A4 Highlighting Hypermethylation and Altered Amygdala Volume in Male Patients With Schizophrenia. Schizophrenia Bulletin. doi:10.1093/schbul/sbaa075
[Publication URL]
https://academic.oup.com/schizophreniabulletin/advance-article/doi/10.1093/schbul/sbaa075/5859552
[Fund(s)]
1) Japan Society for the Promotion of Science (JSPS) KAKENHI, 2) Japan Science and Technology Agency (JST), 3) Japan Agency for Medical Research and Development (AMED)
[Numbers]
1) JP16H06395, JP16H06399, JP16K21720, JP17H01573, JP18H02753, JP18H05428, JP18H05430, JP18H05435, JP18K07567, JP19H03579, JP19K17056, 2) JPMJPR16F8, 3) JP18dm0207006, JP19dm0107097, JP19dm0107120, JP19dm0107123, JP19dm0207069, JP19dm0207074, JP19dm0307001, JP19dm0307002, JP19dm0307004, JP19km0405201, JP19km0405208
[Media 1]
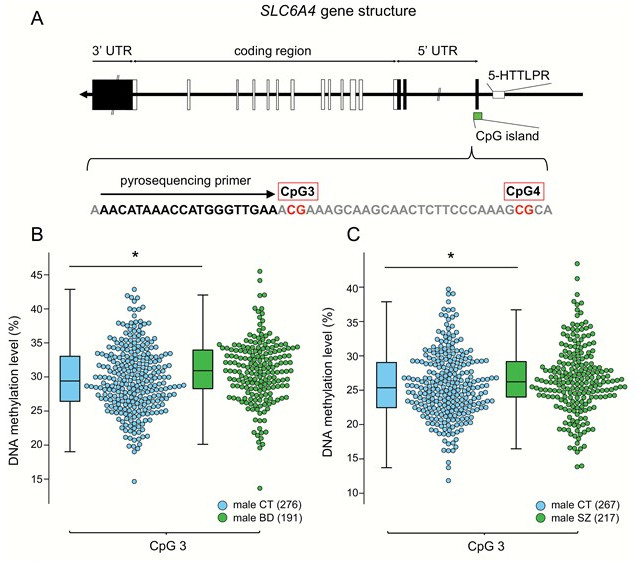
Change in DNA methylation of the SERT gene
A) CpG site where the structure and methylation of the SERT gene were analyzed. Transcription is performed from right to left in the figure, and 5-HTTLPR polymorphism exists in the promoter (transcription initiation) region. Boxes indicate exon regions, black indicates a part that does not encode a protein, and white indicates an exon region that encodes a protein.
B) Comparison of methylation in CpG3 between male bipolar disorder patients (male BD) and healthy male subjects (male CT)
C) Comparison of CpG3 methylation in male schizophrenia (male SZ) and male CT
[Images Details]
CREDIT:
Professor Kazuya Iwamoto
USAGE RESTRICTIONS:
Adapted from figure 1 (A, B, C) of Ikegame, T. et al, 2020. Please note that parts D & E are not included here.
This image is made available under Oxford University Press "Rights retained by ALL Oxford Journal authors". Any further distribution of this work must follow OUP’s terms of reuse.
[Media 2]
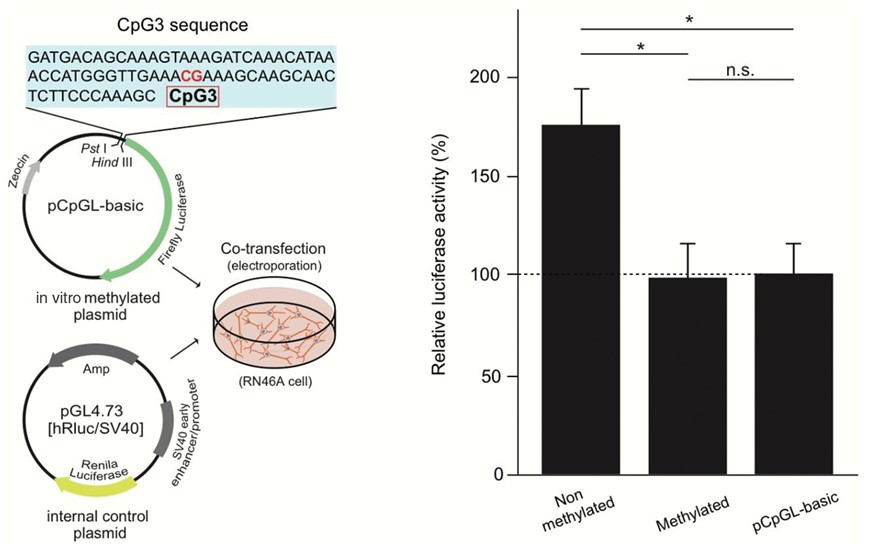
Functional analysis of CpG3 methylation by reporter assay
A) The transcriptional activation ability is measured by introducing a fluorescently labeled DNA chain (reporter construct) artificially methylated at the CpG site into a neural cell line and measuring the activity of the photoprotein.
B) When it is methylated, the transcription activation ability is lost.
[Images Details]
CREDIT:
Professor Kazuya Iwamoto
USAGE RESTRICTIONS:
Adapted from figure 2 of Ikegame, T. et al, 2020.
This image is made available under Oxford University Press "Rights retained by ALL Oxford Journal authors". Any further distribution of this work must follow OUP’s terms of reuse.
[Media 3]
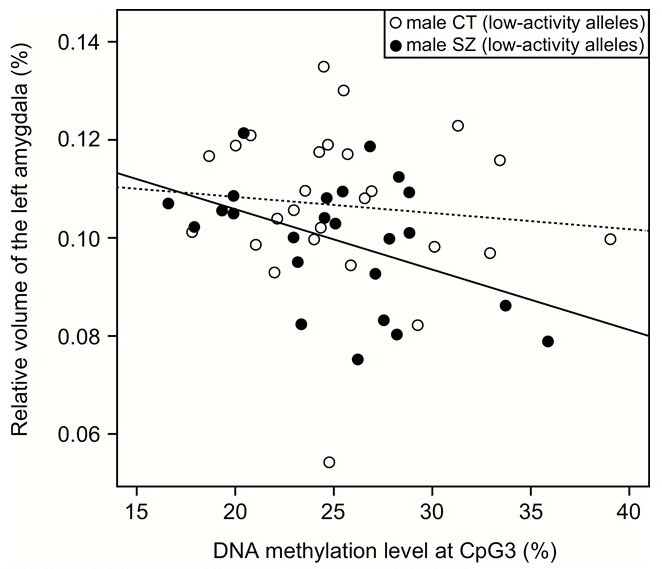
Relationship between DNA methylation rate of CpG3 and left amygdala volume
Left amygdala volume and CpG3 methylation rate are inversely correlated in schizophrenic patients with hypoactive 5-HTTLPR. A significant correlation was found in schizophrenia (SZ) patients (solid line), but not found in controls (CT, dotted line).
Vertical axis: ratio of left amygdala volume to the whole brain (%), Horizontal axis: CpG3 methylation rate (%)
[Images Details]
CREDIT:
Professor Kazuya Iwamoto
USAGE RESTRICTIONS:
Adapted from figure 3 of Ikegame, T. et al, 2020.
This image is made available under Oxford University Press "Rights retained by ALL Oxford Journal authors". Any further distribution of this work must follow OUP’s terms of reuse.
