Microfilter device capable of detecting trace amounts of cancer cells in one mL of blood

A microfilter device that can easily separate and capture trace amounts of cancer cells in blood has been developed by a Kumamoto University research group. The palm-sized device is expected to contribute to the development of new cancer diagnostic technologies based on cancer cells in the blood, such as early detection by blood test, postoperative management, and recurrence monitoring.The blood of people with cancer contains trace amounts of cancer cells (CTCs) that have detached from the primary cancer site. However, the amount of these cells is only a few per milliliter, whereas red or white blood cells number in the billions, making it very difficult to separate and detect them. Although devices for detecting CTCs have been developed in the past, they require expensive equipment and reagents, which has been a bottleneck for their practical application.
The unique microfilter device developed by the Kumamoto University research group can easily and inexpensively separate and capture CTCs without any large equipment. The device is dynamically and three-dimensionally deformed by the fluid force when blood is pumped through it. It also utilizes nucleic acid aptamers, which bind specifically and firmly to target molecules. This enables both size-selective and affinity-selective separation and capture of tiny cancer cells.
A microfilter device for detecting trace amounts of cancer cells
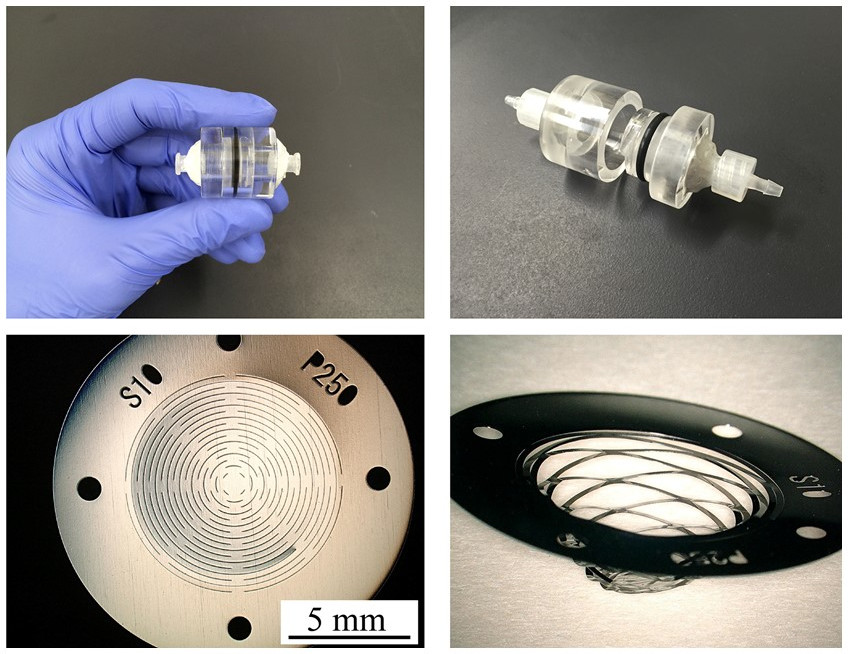
Top: The device is small enough to fit in the palm of your hand.

Bottom left: The device filter is designed with slits allowing for deformation with fluid force.
Bottom right: As blood flows through the device, the fluid force causes the slits to open, changing the shape of the filter and separating blood cells.
Bottom right: As blood flows through the device, the fluid force causes the slits to open, changing the shape of the filter and separating blood cells.
While evaluating the device, researchers demonstrated that it could capture cancer cells even at a concentration of just five cancer cells in one mL of healthy blood. Since there are about five billion (red and white) blood cells in a mL of blood, the device proved to have a very high detection capability. Researchers also found that almost no blood cells were adsorbed by the microfilter, achieving a blood cell removal rate of more than 98%. It also had a high selective detection capability. Furthermore, in a comparative evaluation, they proved that the device was able to detect cancer cells with higher accuracy than existing devices.
Method for detecting trace amounts of cancer cells in blood
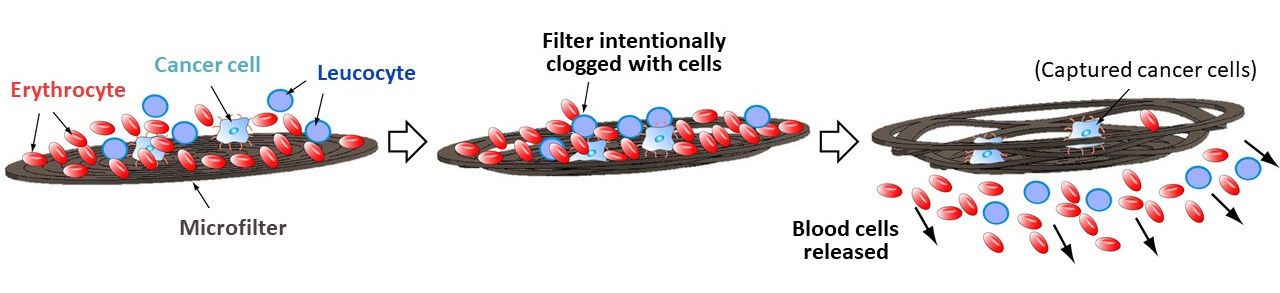
When blood flows over the microfilter, cancer cells in the blood bind to the nucleic acid aptamer. The hydrodynamic force of the blood opens the slits of the filter and the blood cells flow away, leaving the cancer cells on the filter.
"This work demonstrates that our micro-filter device can accurately detect trace amounts of cancer cells in blood," said Associate Professor Yuta Nakashima, who led the study. "We expect it will be adopted for cancer diagnosis and treatment, including for early diagnosis of cancers that cannot be detected by imaging like CT and PET scans, postoperative follow-up, recurrence monitoring, and tailor-made treatments. In the future, we plan to use blood samples donated by cancer patients to verify the practical and clinical application of the method."
This research was posted online in Talanta on 1 June 2021.
[Source]
Fukuyama, S., Kumamoto, S., Nagano, S., Hitotsuya, S., Yasuda, K., Kitamura, Y., ... Nakashima, Y. (2021). Detection of cancer cells in whole blood using a dynamic deformable microfilter and a nucleic acid aptamer. Talanta, 228, 122239. doi:10.1016/j.talanta.2021.122239
[Funds]
- Grant-in-Aid for Young Scientists (A) from MEXT Japan (JP16H06078),
- Grant-in-Aid for Young Scientists (B) from MEXT Japan (JP19H04489),
- Projects to Support the Advancement of Strategic Core Technologies from the Ministry of Economy Japan (17939048)
[Usage Restrictions]
Image credits belong to Associate Professor Yuta Nakashima unless otherwise indicated in the figure caption. To use any of the media contained within this release elsewhere, a reference to the original work (this release) should be included and other restrictions must be followed as indicated.

