DNA methylation changes and characteristics in neurons of bipolar disorder patients

A research collaboration based in Kumamoto University, Japan has revealed the DNA methylation status of gene transcriptional regulatory regions in the frontal lobes of patients with bipolar disorder (BD). The regions with altered DNA methylation status were significantly enriched in genomic regions which were reported to be genetically related to BD. These findings are expected to advance the understanding of the pathogenesis of BD and the development of therapeutic drugs targeting epigenetic conditions.BD is a mental disorder that affects about 1% of the population and requires long-term treatment. Epidemiological studies have suggested that the onset of the disease is related to a complex interaction of genetic and environmental factors. Epigenetic changes in the genome, particularly in the repression of gene expression through DNA methylation, are thought to be deeply involved in the pathogenesis of various diseases, including mental disorders.
Previous studies analyzed DNA methylation using peripheral blood and saliva samples to identify genes with altered DNA methylation and use them as biomarkers. Since mental disorders are diseases of the nervous system, research using brain tissue is particularly important. However, in addition to the scarcity of samples, brain tissue is a mixture of various cell types, such as neurons and glial cells, and is affected by differences in the ratio of cell types contained in the tissue, making accurate analysis difficult.
The researchers used neuronal nuclear markers to fractionate frontal lobe samples from 34 BD patients and 35 healthy subjects into neuronal and non-neuronal cell nuclei. Genomic DNA extracted from each of these samples was then used to examine the DNA methylation status of the regulatory regions of gene transcription using an array method. They found that many genes in both neurons and non-neurons were hypomethylated in BD patients compared to healthy subjects. On the other hand, genes important for psychiatric and neurological functions were found to be hypermethylated in neurons.
To investigate the effects of mood stabilizers—drugs used to treat BD—the researchers cultured human neuroblastoma cells in the presence of mood stabilizers (lithium, valproate and carbamazepine) in effective blood concentration ranges and measured DNA methylation status. They found that about 30% of the regions with altered DNA methylation status in BD patients overlapped with mood stabilizers-induced DNA methylation changes. Most of the DNA methylation changes were in the opposite direction of those in the postmortem brain, which the researchers thought reflected the effects of the treatment. They also measured the expression levels of 10 genes associated with DNA methylation changes and found that the DNMT3B gene, a DNA methyltransferase, was elevated in BD patients, suggesting that it may be associated with neuron-specific DNA methylation changes.
Finally, the researchers compared the genomic regions associated with mental disorders as identified in genome-wide association studies (GWAS) with regions having an altered DNA methylation status. Significant accumulation was observed in the genomic regions reported in the GWAS for BD, while no accumulation was observed in the genomic regions reported for depression and schizophrenia.
"The clarification of neuron-specific DNA methylation changes and characteristics is expected to greatly advance our understanding of the pathogenesis of bipolar disorder," said Professor Kazuya Iwamoto, who led this study. "We also expect that this will help in the development of therapeutic drugs targeting epigenetic conditions."
This research was posted online in "Molecular Psychiatry" on 20 April 2021.
[Source]
Bundo, M., Ueda, J., Nakachi, Y., Kasai, K., Kato, T., & Iwamoto, K. (2021). Decreased DNA methylation at promoters and gene-specific neuronal hypermethylation in the prefrontal cortex of patients with bipolar disorder. Molecular Psychiatry. doi:10.1038/s41380-021-01079-0
[Funding]
1) KAKENHI Grants from MEXT Japan (16H06395, 16H06399, 18H05435, 16K21720, 18H05428, 18H02753, 18H05430, 18K07567),
2) Grants-in-Aid for Challenging Exploratory Research from MEXT Japan (JP15gm0510002, JP20dm0307001, JP20dm0307004, JP20dm0207069, JP20dm0107123, JP20dm0207074, JP20km0405208)
[Image 1]
Hypomethylation in transcriptional regulatory regions and hypermethylation of genes important for neural function
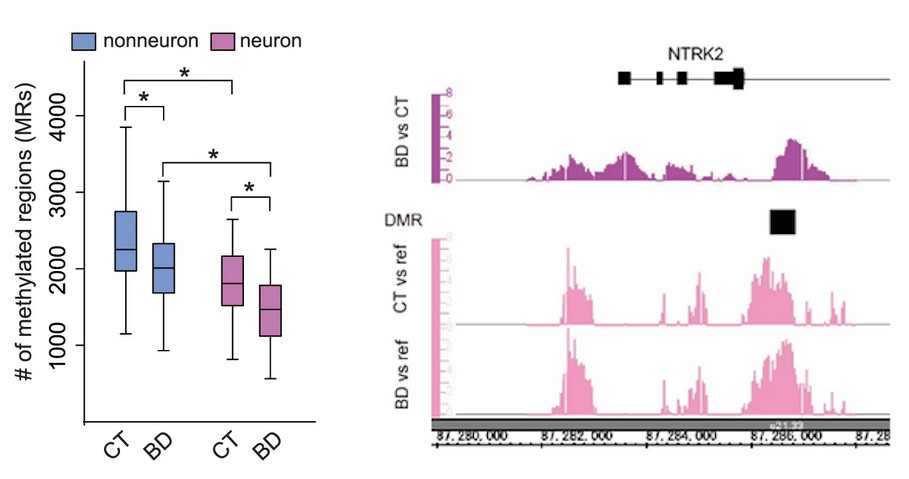
(Left) The number of hypermethylated DNA regions in both neuronal and non-neuronal cells is reduced in the bipolar patient group (BD) compared to the healthy group (CT).
(Right) Methylation status of the BDNF (brain-derived neurotrophic factor) receptor gene (NTRK2), which is important for mental and neurological functions. The upper panel shows the hypermethylated region in patients, and the lower panel shows the DNA methylation status in patients and healthy controls.
[Credit]
Professor Kazuya Iwamoto
[Usage Restrictions]
Left: Fig. 1(b), Right: Fig. 2(d) of Bundo, M. et.al., Molecular Psychiatry, 2021.
This image is made available under the terms of CC by 4.0. Any further distribution of this work must follow the terms of this license.
[Image 2]
Accumulation of genes with differential DNA methylation status in neurons in GWAS regions
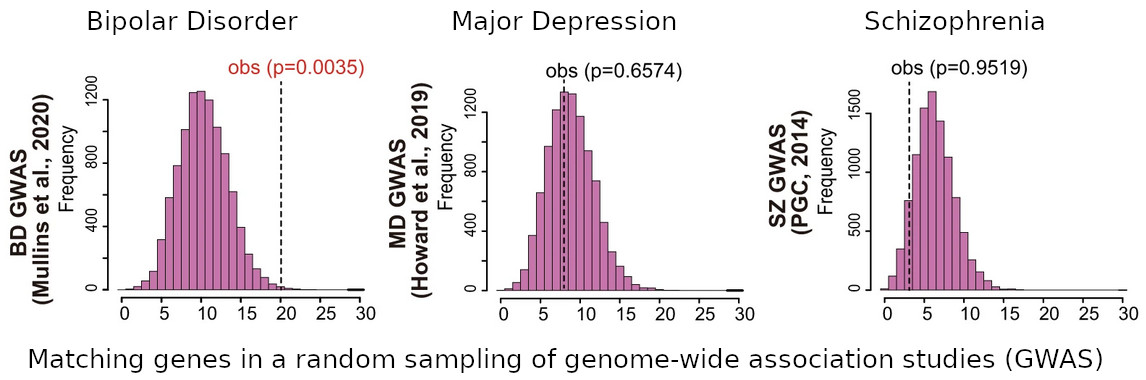
Researchers performed random sampling of the same number of gene transcriptional control regions in the genome that showed DNA methylation variation 10,000 times to determine the number of matches with GWAS regions for bipolar disorder, major depression and schizophrenia, and to obtain p-values for the measured values (obs). A significant accumulation was found in the GWAS for bipolar disorder, while no significant accumulation was found for major depression or schizophrenia.
For each disease, the following data were used.
Bipolar disorder: data from Mullins et al, 2020.
Major depression: data reported in Howard et al, 2019.
Schizophrenia: data from Psychiatric Genomics Consortium, 2014.
[Credit]
Professor Kazuya Iwamoto
[Usage Restrictions]
From Fig. 4(c) – column 1, rows 4, 1, & 2 respectively of Bundo, M. et.al., Molecular Psychiatry, 2021.
This image is made available under the terms of CC by 4.0. Any further distribution of this work must follow the terms of this license.

