New bioresources for plant peptide hormones using gene editing technology
Peptide hormones are important growth regulators that play various roles in many organisms. Although the importance of genes that encode peptide hormones is broadly recognized, most peptides have yet to be functionally characterized. Using genome editing technology, researchers from Kumamoto University, Japan generated a new, comprehensive genetic resource collection of plants with knocked out CLE peptides-encoding genes. CLE peptides are a group of plant-specific peptide hormones that play a role in cell signaling and are regulated by CLE genes. This open collection is expected to contribute to future studies on how peptide hormones work in plants.Genome editing, a technology that enables us to knockout or modify specific genes, has been in the spotlight recently. The "CRISPR/Cas9" method was established in 2013 with successful cases of genome editing reported in animals, yeasts, plants, and many other organisms. Before CRISPR/Cas9, genome editing required complicated procedures and was thus not popular in research that targeted multiple genes. The establishment of CRISPR/Cas9, on the other hand, brought great advances to genome editing due to its high efficiency, high target specificity, and simplicity. Because of these advantages, researchers gained a way to investigate the function of genes, such as analyzing the impact of knocking out specific genes on the behavior of organisms and/or cells.
Recent advances in plant science have discovered various areas where peptide hormones provide valuable information in plant life cycles, for example determining the number of cells and size of tissues, controlling pollination, and responding to climate change or disease. Many scientists are currently researching plant genetics because the genes for many uncharacterized peptide hormones have not yet been discovered. Even though peptide hormone-encoding genes are difficult to study, they can generate a wealth of genetic resources for future research.
Using CRISPR/Cas9, the Kumamoto University research team attempted to accelerate plant peptide hormone research. They selected Arabidopsis thaliana, a model plant with 32 CLE peptide-encoding genes, and generated a collection of gene knockout tools that corresponded to each of the 32 genes. By establishing these mutant Arabidopsis plant lines, they made it possible to more easily investigate the function of each gene.
To evaluate the effectiveness of the gene knockout, the research team looked at CLV3, the most studied CLE peptide. It is known to have a role in suppressing cell divisions at the growing points of stems. As expected, the CLV3 mutant of Arabidopsis plants produced irregularly shaped fruit due to an increased number of cells.
Furthermore, the researchers were able to use the mutant plants to make what is believed to be the first report on the biological relevance of gene CLE44. Although it was expected that CLE44 regulates the number of cells in the vascular bundle, research into that hypothesis had not been performed due to insufficient bioresources. With the help of their newly developed mutant plant lines, the Kumamoto University researchers observed a decrease in the number of vascular bundle cells in the CLE44 mutant plants, thereby confirming its role in vascular development.
"Our genetic resource collection has the potential to help study unknown peptide hormones like CLE44," said Assistant Professor Takashi Ishida of Kumamoto University. "We hope this approach becomes a model for similar research in the future."
This research was posted online in "Plant and Cell Physiology" on 25 September 2017 and will be featured as a “Research Highlight” in an upcoming volume of the journal.
[Resource]
Yamaguchi, Y. L., Ishida, T., Yoshimura, M., Imamura, Y., Shimaoka, C., & Sawa, S. (2017). A collection of mutants for CLE-peptide-encoding genes in Arabidopsis generated by CRISPR/Cas9 mediated gene targeting. Plant and Cell Physiology. doi:10.1093/pcp/pcx139
[Paper Info]
TITLE: A collection of mutants for CLE-peptide-encoding genes in Arabidopsis generated by CRISPR/Cas9 mediated gene targeting
AUTHORS: Yasuka L. Yamaguchi, Takashi Ishida, Mika Yoshimura, Yuko Imamura, Shinichiro Sawa
JOURNAL: Plant and Cell Physiology
DOI: 10.1093/pcp/pcx139
URL: https://academic.oup.com/pcp/article/4222595/A-collection-of-mutants-for-CLE-peptide-encoding
[Funds]
Grant-in-Aid for Scientific Research from the Japanese Society for the Promotion of Science
[grant numbers JP26440151, JP14J40052, JP25440134, JP25119713, JP24114009, JP24370024]
[image1]
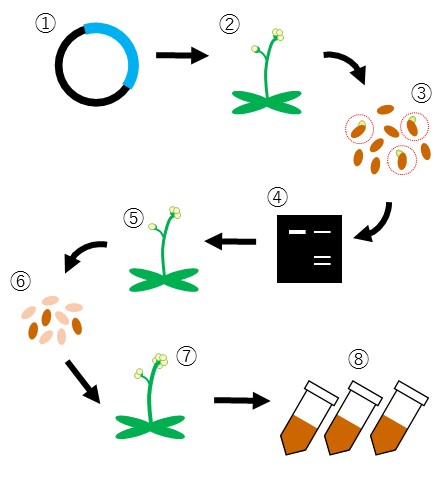
Schematic representation of procedures for the generation of gene disruption lines.
CAPTION:
- Construction of gene targeting vectors
- Plant transformation
- First generation plant selection
- Gene disruption efficiency evaluation
- Onset of genome editing events
- Selection of plants that lost gene disruption tools (preventing unpredictable/extra genome editing)
- Selection of plants containing target gene mutation
- Establish mutant line/publish collection
[image2]
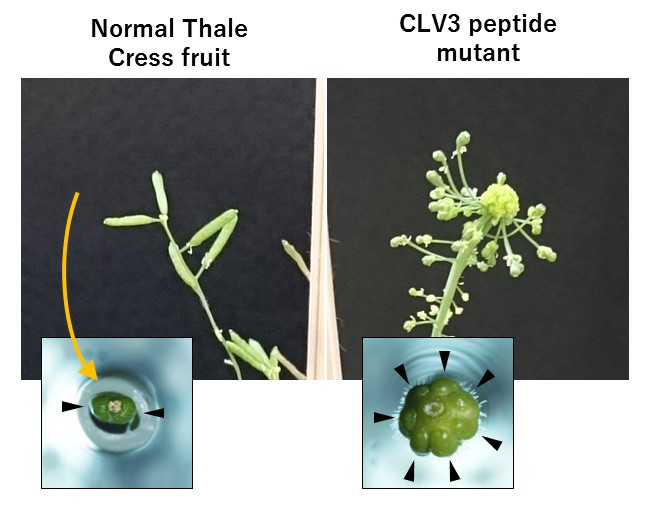
Fruit shapes in normal Thale Cress plants and CLV3 mutant Thale Cress plants.
CAPTION:
In Arabidopsis mutants lacking the CLV3 peptide, the amount of fruit tissue is increased.
[image3]
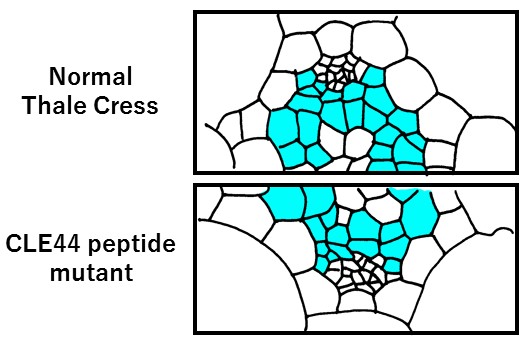
Representation of CLE44 mutant microscopic observations
CAPTION:
Procambial cells regulated by CLE44 are shown in light blue. In the CLE44 peptide mutant, the number of cells is decreased.
[FOR ALL IMAGES]
Credit:
Assistant Professor Takashi Ishida
Usage Restrictions:
This image may only be used in conjunction with the accompanying release, or stories written about the work described in the release with reference to the original work.

